|
Defining heavy labeled residues.
Working with isotopes, e.g. heavy labeled amino acid residues, is fairly straightforward in GPMAW. However, you need to do a few preparations in order to make it function smoothly.
In the following we want to be able to quickly change from a standard mass presentation to a situation where all lysine residues have been replaced with heavy lysines, i.e. 12C replaced with 13C and 14N with 15N.
To begin with we have to define the 'new' atoms in the atom table. Open the Edit | Edit mass dialog box and select the 'Atomic weights' tab.
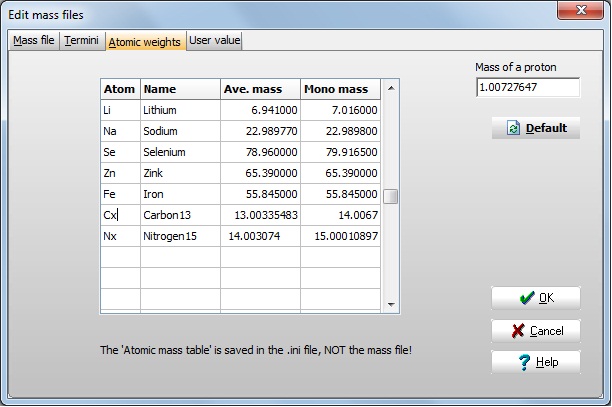
Move to the bottom of the table and enter names and mass values for 13C and 15N. Remember that the Atom name can only be two characters long, and has to be different from all other atoms in the table.
Select OK and re-open the Edit | Edit mass file, now on the first 'Mass file' tab.
Edit the compositions of Lys and Arg as indicated in the figure below (e.g. Lys from C6H12N2O2 to Cx6H12Nx2O2)
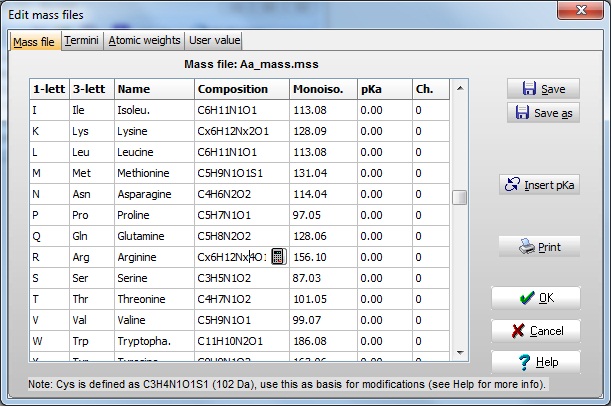
Now select Save as and enter a new name for the mass table, e.g. 'HeavyLysArg', and select OK. You can now in your drop-down mass file selector find the new mass file.
If you perform a quick check on your currently opened sequence, you can see that you have a new mass calculation:
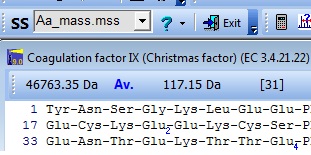
Changing the mass file from aa_mass to HeavyLysArg changes the average mass of the protein from 46763 Da to 46971 Da, reflecting the increased mas of the heavy atoms.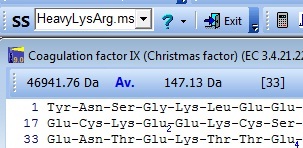
Note: If you are working with differently modified residues (e.g. free and alkylated Cys), you have to make a mass file for each of the combinations.
If you just want to compare mass values of the light and heavy peptides, you can get the peptide list to display them easily, check here.
|