|
Chapter B – Calculating mass values.
1 – The mass file.
Calculating mass values is central to handling sequences in GPMAW. As the way mass values are used are quite varied, they are subsequently calculated in a flexible (although a little complicated) way.
Peptide/protein mass values are calculated based on ‘mass files’. These are constructed of residues. Each residue contains: 1- and 3-letter code, name of residue, atom composition, pKa and charge of side chain. The residue is defined by the 1-letter code, which is what the sequence is stored as in memory/on disk.
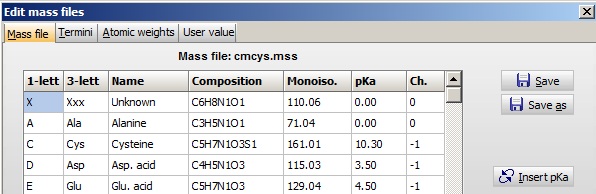
The table is edited through the menu Edit | Edit mass file where you have tables for the amino acid residues, peptide termini and atom mass values.
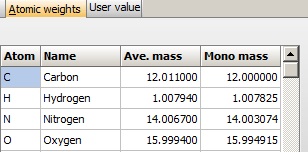 Atom mass table: The basis of the mass calculations is the composition, which again is based the table of atom mass values. By default this list contains the monoisotopic and average mass value of the 16 most common atoms, but it can be extended to 24 values. Atom mass table: The basis of the mass calculations is the composition, which again is based the table of atom mass values. By default this list contains the monoisotopic and average mass value of the 16 most common atoms, but it can be extended to 24 values.
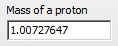
Note that the mass of a proton is defined separately.
The mass file: The mass files are loaded upon startup of the program and form the basis for calculating peptide and protein sequence mass values.
By default GPMAW is delivered with a number of files, which only differ in the compositions (mass value) of cysteine. The currently loaded mass file is displayed in the main toolbar next to the ‘SS’ button  . The default file is called AA_mass.MSS. If you change the active mass file by selecting a new in the drop-down box (i.e. click on the arrow and select), you will change the mass value of all proteins and peptides opened, to the values defined in the new file. Note: this is the easiest way of changing the mass of all residues of a given kind. . The default file is called AA_mass.MSS. If you change the active mass file by selecting a new in the drop-down box (i.e. click on the arrow and select), you will change the mass value of all proteins and peptides opened, to the values defined in the new file. Note: this is the easiest way of changing the mass of all residues of a given kind.
Please note that Cysteine is calculated differently from all other amino acids. If you define Cys as being in the reduced state, this will be changed by the program to the oxidized state (i.e. a mass value of 103 will be changed to 102). The oxidation step of Cys is controlled through the ‘SS’ button. When the button states ‘SS’, Cys will be calculated as 102 Da, when the button is pressed and state ‘SH’, Cys will be calculated as 103 Da. If Cys is defined as something else but 102/103 Da in the current mass file, the SS button will be inactive.
The most typical example for the use of a mass file is when you alkylate a protein. E.g. if you reduce and alkylate your cysteines with iodoacetamide, you will just change the mass file from AA_mass.MSS to acetamide.MSS, and all your mass values will be based on Cys = 161 Da.
Termini: This is a separate table from mass files, which specifies the status of the N- and C-termini. By clicking on the termini buttons in the sequence window  , you will open a dialog box enabling you to select the chemical status of the termini. The termini are edited in the same dialog box as above. , you will open a dialog box enabling you to select the chemical status of the termini. The termini are edited in the same dialog box as above.
2 – Other ways of changing the mass.
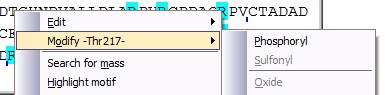 In addition to changing the mass of a residue in a global way as described above, you can modify each residue individually. If you right-click on a protein residue, the pop-up menu displays a ‘Modify –Xxx-‘ option, which have a sub-menu detailing the ‘simple’ modifications available for the particular residue. Selecting one of these will put the modification into the sequence table and the mass value will be recalculated. Whenever a residue has been modified, it will be displayed in red in the sequence. In addition to changing the mass of a residue in a global way as described above, you can modify each residue individually. If you right-click on a protein residue, the pop-up menu displays a ‘Modify –Xxx-‘ option, which have a sub-menu detailing the ‘simple’ modifications available for the particular residue. Selecting one of these will put the modification into the sequence table and the mass value will be recalculated. Whenever a residue has been modified, it will be displayed in red in the sequence.
Note: the modification will be carried on into the peptide window and several other.
When you have modified a sequence, you need to save it to ‘keep’ the modification.
If you ‘un-check’ the ‘Strict modification check’ at the bottom of the menu, you can select any of the ‘simple’ modifications for any residue (although it may not have any biological or chemical relevance).
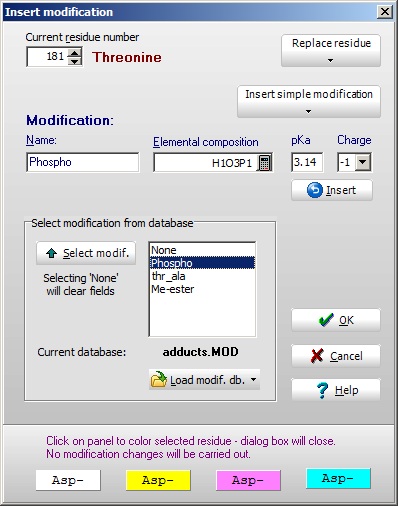 You may also double-click on a particular residue to bring up the ‘Insert modification’ box. In the top left-hand box you will see the residue and number, which can be changed. Two drop-down buttons to the right will enable you to either replace a residue or insert a ‘simple’ modification (the same menu as above). You may also double-click on a particular residue to bring up the ‘Insert modification’ box. In the top left-hand box you will see the residue and number, which can be changed. Two drop-down buttons to the right will enable you to either replace a residue or insert a ‘simple’ modification (the same menu as above).
In the central box is listed the available modifications in the currently loaded modification file (see below). You can either double-click or use the ‘Select’ button transfer values to the edit box above. These can also be filled directly.
Selecting ‘OK’ will transfer the information to the selected residue in the sequence.
The four bottom panels will just transfer the corresponding color as a background to the residue.
The modification file
This is a file which contains up to 30 modifications specific to one or more residues. You may have as many files as you like, but only one can be loaded at any given time. In addition to the ‘Insert modification’ box, the modification file is used in a number of other functions (e.g. mass searches).
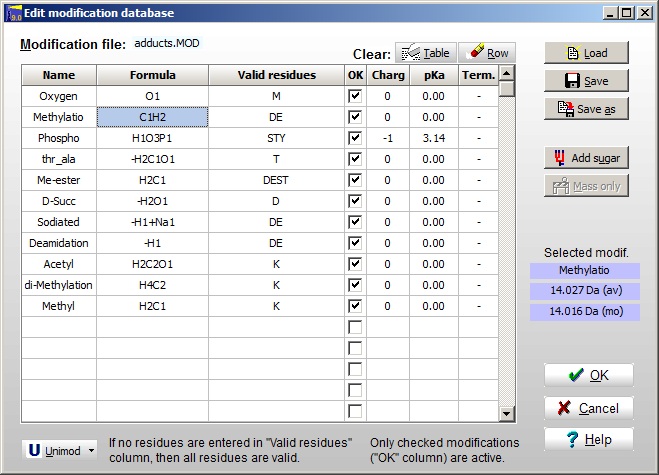
The modification file is edited through Edit | Edit modification file. For each modification you have to edit the Name, the Formula, Valid residues (if none are specified, all are valid), the OK box enables you to enable just some to the entries. The Charge, pKa and Terminal are optional. The mass of the currently selected entry is shown to the right. The Unimod file can be opened at the bottom, and you can select entries to the modification file by double-clicking on the Unimod entries. If you do not know the chemical composition of a modification, you can enter it by selecting an empty line and click the ‘Mass only’ button.
A modification file has to be saved to disk before you can use it in GPMAW.
|