|
|
|
|
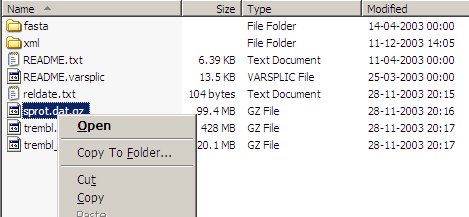
The Swiss-Prot is the best annotated protein database and as such an absolute requirement in the toolbox of any protein chemist. The database is maintained by SIB and EBI and details can be found at the Expasy web site. Note: Although the database can be freely downloaded, it is not free-ware. Commercial users need to obtain a license. Non-commercial uses (e.g. universities and other educational institutions) are free. Start by downloading the database from the EBI FTP site (ftp://ftp.ebi.ac.uk/pub/databases/sp_tr_nrdb/) and copy it to its own directory (note that the total space requirement may easily be in the order of 6-700 MB). Using the Internet Explorer from Windows, it looks like this:
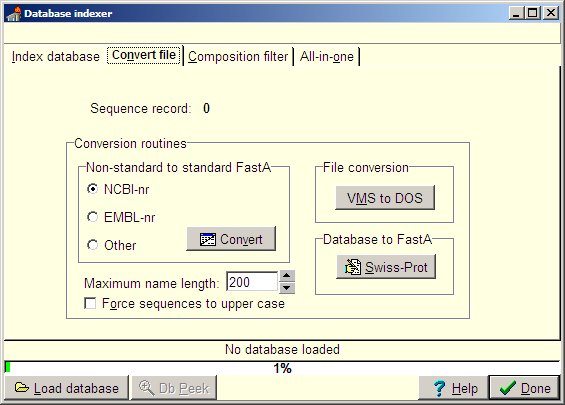
Right-click on the sprot.dat.gz file, select “Copy To Folder...” and in the following “Save as” dialog box select an appropriate directory. Decompress the ‘sprot.dat.gz’ file using an appropriate tool like gzip, WinZip or Zipmagic, and create the ‘sprot.dat’ file. Once the file has been decompressed, the .gz file can be deleted (thus saving ~100 MB of disk space). Start DBindex (either directly or through GPMAW - Utilities|Call DBindex). In the opening dialog select the “Convert” button - this takes you to the “convert” page of the dialog.
As the file has been downloaded from a Unix based server, you need to convert it to DOS format: Click on the “VMS to DOS” button and select the ‘sprot.dat’ file in the ‘File open’ dialog box. Upon selection, the file is converted in ½-1 minute. The converted file replaces the old one. Select the “Swiss-Prot” button to create a FastA-formatted version of the file. A dialog box asks for database type, and selecting Swiss-Prot gives the FastA name of “SWISS”. This name can be edited. Selecting ‘OK’ creates the FastA database in ½-1 minute. Two files are created, SWISS.SEQ (the FastA database) and SWISS.IDX (the index from the FastA database into the main Swiss-Prot database - sprot.dat). From here you can perform various indexing of the FastA version of the database. The indexing function on the first page of the database indexer generates indices for text searching from within GPMAW. |
|||||
|
Site last updated: February 14, 2025 |
|||||