|
|
|
|
D – Post-translational modifications. 1 - Obtaining the sequence – Swiss-Prot. This time we will retrieve a sequence from the Swiss-Prot database. The Swiss-Prot/UniProt database is supplied on the original GPMAW installation CD-ROM. The database cannot be accessed directly on the CD-ROM so you have to copy it to the hard drive using the installation program (can be done after the main installation of GPMAW). If you do not have the CD-ROM, or if you need a more recent version of the database, you can download it from the Internet (e.g. ftp.ncbi.nlm.nih.gov, ftp.ebi.ac.uk, ftp.expasy.ch), but you have to convert it to FastA format and index it using the Dbindex utility before searching the database.
Select File|Open Database| FastA (or click the FastA button in the main toolbar) and the search database dialog box (right) will open. The first time you open the dialog it will be empty and you will have to navigate to the relevant database file (with a .trg extension) in order to get access to it. Once accessed, the program ‘remembers’ the database (or rather the 5 most recently accessed), and you can open it directly by either pressing the relevant button in the bottom left corner of the dialog (the buttons show the first two characters of the database name, the fly-by help shows the full name), or you can access it at the bottom of the File menu item. After you have installed the Swiss-Prot database from the CD-ROM, press the ‘Open database’ button, and in the ‘Open’ dialog you navigate to the Swiss-Prot database. When you have found the correct database directory, you select the file ‘swiss.trg’. When the ‘Search database’ dialog opens, you are greeted with information on the database (see above). You can see the selected database in the title (Swiss) and in the status line at the bottom of the dialog where the complete path to the database is displayed. On future access to a FastA search, you can select File| Open FastA database in the main menu or the buttons at the bottom of the dialog box. The fastest way to retrieve a sequence is by using the accession number (if known). Just enter the accession number (e.g. P23805) in the ‘Accession #’ field, press the search button and the name of the protein will show in the result box:
Highlight the name and press the ‘Retrieve’ button and the sequence will be imported into GPMAW as a sequence window. If the ‘Retrieve annotation’ tick-box is selected (it is ‘on’ by default), the entire database entry will be copied into the annotation page of the sequence window. If you do not know the accession number, you will have to search on the basis of the protein name and, perhaps, species. Please remember that you are searching the FastA formatted version of the database, not the entire database (even though you will retrieve the full database entry). You are thus limited to words that are present in the name line of the database entry. This line will usually also hold the species name. If you need to search in other parts of a database entry, you will need use one of the web search engines (e.g. the EBI, www.ebi.ac.uk, or Expasy, www.expasy.ch for the Swiss-Prot database).
You could also have entered ‘factor’, but the search returns so few entries that it does not matter. Normally you use ‘and’ for the search parameters (in this case ‘coagulation and bovine’), but you can also use ‘or’ (works only reliably for two parameters). If you get an ‘I/O error 87’ your search terms are too loose and you have to narrow them. You should always put the most selective term first (e.g. if you use ‘human’ make it the last term).
Select ‘Done’ when you are finished retrieving sequences. Save the sequence(s) to disk (remember you can save multiple sequences to the same file). 2 – Inserting post-translational modifications If you have retrieved your sequence from a Swiss-Prot database as illustrated above you should have a green ‘a’ in the sequence toolbar If the ‘a’ is not green and there is no information in the annotation page, you should go back and make sure that the ‘Retrieve annotation’ tick-box is ticked in the ‘Search database’ dialog (alternatively there was no annotation to retrieve from the database, either because the full database was missing or the cross-index file was not functional). If you still do not get the annotation, your sequence database is not set up correctly and you should reinstall it (e.g. by using the install program on the CD-ROM or re-index a downloaded database). Note that you will not get an annotation when you use other databases like the EMBL-nr or NCBI-nr as these databases are originally in FastA format and does not contain other information than name, accession number and sequence. Click on the green ‘a’ in the sequence window toolbar. Looking at the secondary modifications in the annotation, they should look like this for human Factor X (2 coloums) FT SIGNAL 1 ? As this is a Swiss-Prot annotation, GPMAW has two was of inserting the modifications into the sequence, a manual and a semi-automatic one. As the manual one is generally applicable, it will be presented first. Manual method for posttranslational modifications: When you want to add post-translational modifications to your sequence, you can either add these as a ‘new’ residue or as an add-on modification (see below). If you have a large number of residues and/or you analyze this residue regularly, you should use ‘New residue’. If you on the other hand only have a few modifications of a given kind, you should use ‘Add-on modification’.
The first task is then to create a ‘new’ amino acid residue in GPMAW. From the main menu select Edit|Edit mass file. Scroll the list of residues until you come to the ‘Unknown’ section of the mass list. As all 1-letter codes have to be unique (and not a punctuation mark, ‘-‘ or ‘$’) select the 1-letter code ‘U’. The default for ‘unknown’ residues is that the 3-letter code is a triplet of the 1-letter code, but as it only has to be unique, so changed the 3-letter code to ‘Gla’, and the name to CarboxyGlu. The amino acid composition you copy from Glu (C5H7N1O3) and paste it into the composition field. Then double-click on the composition to invoke the ‘Elemental composition’ editor and increased the number of ‘C’ atoms by one, and the number of ‘O’ by two. Alternatively you can just edit the field directly (click twice or select and press F2). Save the file using the ‘Save’ button. If you only want the modification for special occasions, you can use the ‘Save as’ button and give it a unique name. You can then select the file through the mass file selection box in the main toolbar. Return to the sequence window and start editing (Edit|Edit sequence). Start by changing the 12 Glu residues (res. 46, 47 … 79) in the N-terminal to ‘U’ (our new carboxyglu). Remember that the ‘Cursor’ field in the sequence editor reports the residue number before the cursor. ‘Add-on’ modification: The modifications of individual residues can either be carried out from the sequence editor or from the sequence window:
The panels at the bottom of the dialog box will color the residue in the sequence window using the displayed background color (and the dialog will close immediately). The residues in position 208 and 485 are O-glycosylated and 218 is N-glycosylated. The actual carbohydrate groups are not mentioned in the annotation, and you will have to refer to the main literature to determine the actual modification to enter in the sequence. N-linked glycosylations can be quite heterogeneous, and identifying the exact glycosylation pattern can be difficult – see the glycosylation tool in section C.3 below. Now you can enter the disulfide links by clicking the ‘Cross-links’ button as described for insulin in the previous example. In the sequence editor you can now insert a cleavage (the dash character ‘-‘) after residue 233 (the activation peptide) and remove the initial 40 residues (the signal- and the pro-peptide). Click on ‘OK’ and you have your final edited sequence:
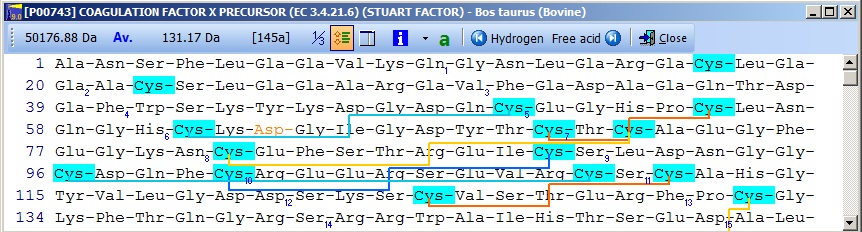
Notice that the Cys residues have been highlighted in order to better locate them. The Glu residues in the N-terminal region are correctly labeled as Gla. The modified Asp residue is red (if you move the mouse cursor over it you can see the actual modification in the top right) and the cross-links are displayed as red lines. If you click on the ‘SS’ button in the main toolbar, you will ‘reduce’ the cysteines (the mass of each Cys will increase by one Da) and the cross-links will be grayed. Semi-automatic method for posttranslational modifications: If you have imported a Swiss-Prot record with full annotation, the ‘a’ button on your sequence window will be green. If you press this, the ‘annotation window’ will open showing you the full annotation. This is a two-page window that upon activation of the second page, ‘Feature table’, will present you with the following view:
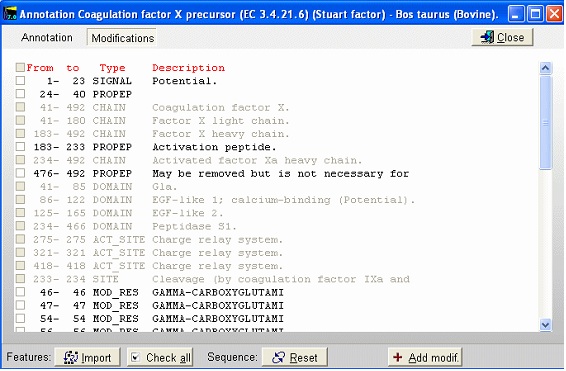
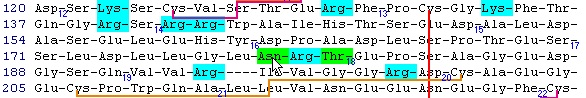
This is the FT (feature) section of the Swiss-Prot record. The parts that are recognized by GPMAW are shown in black, while unrecognized ones are grayed out. You can now check those features that you want to import into the sequence and pressing the “Import” button will close the window and transfer the modifications to the sequence window. If the program is unable to transfer some items a dialog box will inform you. Modified residues will be transferred as individual modifications of residues. Signal and propeptides will be removed from the sequence. Remember to import the residue modifications first and then modifications that change the sequence length. You may include both types in the same ‘import’ session, as GPMAW will do the size modifications last. Once you have changed the size of the protein, GPMAW will not be able to transfer residue specific modifications – in this case you will have to ‘Reset’ the sequence (i.e. remove all modifications and return to the sequence as listed in the annotation). You can add your own modifications through the ‘+ Add modif.’ button. Note that only simple modifications defined in Swiss-Prot (and GPMAW) can be added. Remember to save after making changes. 3 – N-linked glycosylations N-linked glycosylations are in MALDI mass spectra often detected by observing a mass difference of approximately 291 Da between peaks, arising from sialic acid differences between different glycosylation forms (or partial loss of sialic acid in the mass spectrometer). Using factor X protein and doing a tryptic digest you can proceed as follows: From the quick color menu you select ‘Basic residue’ and then ‘N-glycosylation’. You can now easily locate both tryptic cleavage sites and the N-glycosylation sites.
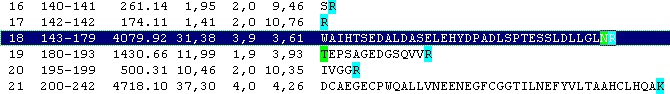
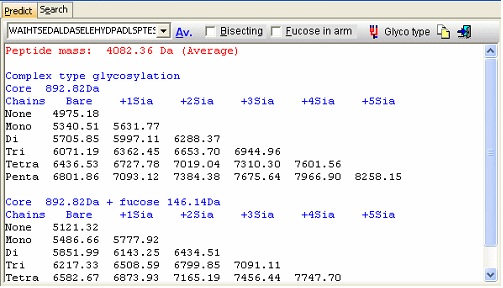
Looking carefully at this sequence you will notice that the identification of this glycosylation may be difficult: Asn178 is located in a fairly large peptide (4082 Da unmodified). The terminating Arg is just after the glycosylated residue and may thus interfere with cleavage. The cleavage before the peptide is a double Arg, which again can lead to heterogeneity (missed cleavages). Taking this into account, you can simulate a cleavage (Cleavage|Automatic digest…). In the digest parameters select a partials level of 2 (= up to two missed cleavages) in order to include the heterogeneity in the resulting peptides. In the resulting peptide box you can easily locate the potential N-glycated peptide as number 18.
Right-click on the peptide and select ‘N-glycosylation’ from the pop-up menu.
In the resulting dialog you now get a mass list of the most common ‘complex type’ glycosylations linked to the peptide in question. Check the ‘Bisecting’ box to add a bisecting GlcNAc or check the ‘Extra fucose’ to add an extra fucose residue to the carbohydrate chain. The ‘Glyco type’ button switches the display to show masses of high mannose structures. If you switch to the ‘Search’ page you can use a mass list (e.g. typically from an ms/ms experiment) to search for valid glycostructures. As the number of structures is astronomical, the search is only carried out for the ‘standard’ types as shown above (+/- fucose) plus and minus any sugar defined in the ‘sugars.mod’ modification file.
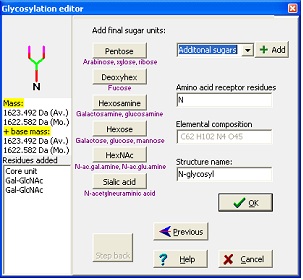
In the wizard, you start by entering your base mass. This is anything attached to the carbohydrate, and can be peptide or derivatization agent. Then you select either N-linked, O-linked or other glycosylation. For the N-linked you then chose the kind of structure to extend a core structure with before ending at the editor where you can add any kind of carbohydrate defined in the ‘Sugars.mod’. Standard sugars can be entered by pressing the appropriate buttons, note that sugars are listed as base type only (e.g. Hexose covers galactose, glucose and mannose) as you cannot distinguish between these isomers using mass spectrometry. As you add residues, the ‘Step back’ button lights up, enabling you to ‘undo’ selections and rebuild your structure.
|
|||||
|
Site last updated: February 14, 2025 |
|||||

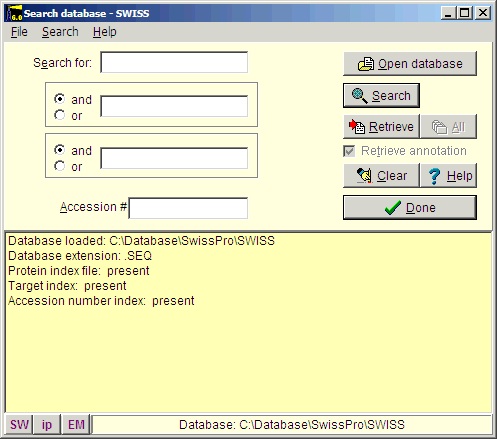 The main reason for using the Swiss-Prot database is that this database is the best-annotated and curated protein database. This also means that the database is less redundant and the sequences found here are more likely to be ‘correct’ than auto-translated sequences from a nucleotide database.
The main reason for using the Swiss-Prot database is that this database is the best-annotated and curated protein database. This also means that the database is less redundant and the sequences found here are more likely to be ‘correct’ than auto-translated sequences from a nucleotide database.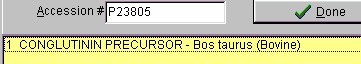
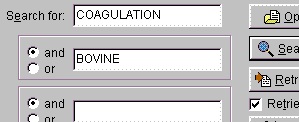
 The results of the above search returned 7 hits. Double-click on ‘factor X’ or highlight and press
The results of the above search returned 7 hits. Double-click on ‘factor X’ or highlight and press 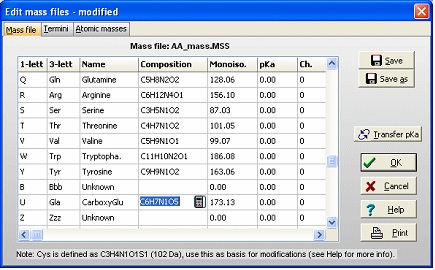
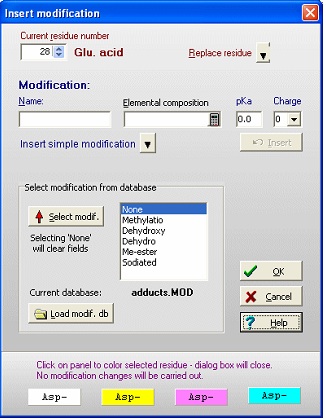 For both 1) and 2) you get the same ‘Insert modification’ dialog box (right). If the modification is in a modification database (in the example the adducts.mod file has been loaded), you can select it from the list box. If not, you enter a name (hydroxylation) and elemental composition (O1 – one extra oxygen) and click
For both 1) and 2) you get the same ‘Insert modification’ dialog box (right). If the modification is in a modification database (in the example the adducts.mod file has been loaded), you can select it from the list box. If not, you enter a name (hydroxylation) and elemental composition (O1 – one extra oxygen) and click