|
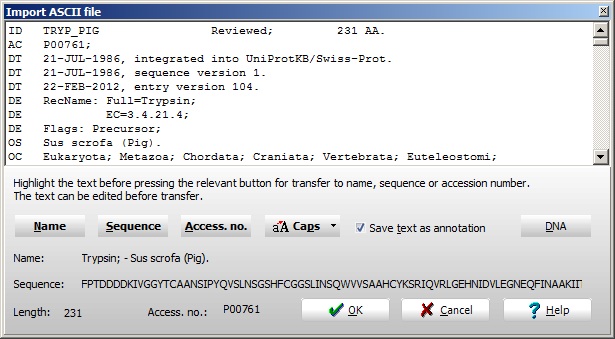
In most cases the file format will be recognized directly and the record will be split into:
Name
Sequence
Accession number
If this automatic parsing is not done, check here how to do it manually.
Click the OK button to retrieve the sequence into GPMAW.
NOTE: Accession numbers starting with O, P, Q are searched through the EBI server and the sequence records are retrieved in Swiss-Prot format. All other accession numbers are searched in the Entrez database (NCBI). The complete sequence record is saved on the annotation page. The Swiss-Prot format is recommended, as GPMAW is able to extract information on secondary modifications through the annotation dialog).
|